Open Source Malaria (OSM) publishes its first paper today. The project was a real thrill, because of the contributors. I’d like to thank them.
Skepticism about open source research is often based on assumptions: that people will be too busy or insufficiently motivated to participate, or that there will be a cacophony of garbage contributions if a project is open to anyone. I’m not sure where such assumptions come from – perhaps people look first for ways that things might fail. We can draw upon many experiences of the open source software movement that would suggest such assumptions are poor. We can draw on successful examples of open collaboration in other areas of science, such as the Human Genome Project and the projects it has spawned, as well as examples in mathematics and astrophysics. This OSM paper addresses open source as applied to drug discovery, i.e. experimental, wet lab science in an area where we normally expect to need secrecy, for patents. It is based on the experience of 4-5 years of work and describes the first series examined by OSM. The paper argues strongly against these assumptions, since:
- Many people contributed enthusiastically
- The contributions came from a wide range of institutions, from Pharma through to Universities, from undergrads through to professors.
- Those contributions, many of which were unsolicited, were of a high quality.

Roll Credits
So who did what? Time for a bit of a credit reel. This is to express sincere thanks to the contributors, but that conveys the wrong idea of ownership. The philosophy behind the project is shared ownership, with a CC-BY license. We all share the project’s outputs, and we all worked together to get there – that’s the whole point. But still, I am keenly grateful that people got on board a highly speculative project at such an early stage.
Pharma as Open Source Partners
Without GSK Tres Cantos (Francisco-Javier Gamo, Félix Calderón, Benigno Crespo, Maria Lafuente-Monasterio) I’m not sure where we’d be. Their still-astonishing paper in 2010, containing thousands of beautiful hits against malaria, kick-started so much. They continued their inputs to the project, from experiments through to invaluable smaller contributions that take time to make, such as revealing that analogs being suggested by the community had/had not been evaluated by GSK.

GSK’s Transformative Open Dataset for Malaria Drug Discovery
This is a powerful reminder that just because the business model of Big Pharma means it cannot officially countenance open source, the companies are filled with immensely talented people who may be free to contribute, and are interested in doing so. Sanctioned freedom to operate is an enlightened policy and could be a real driver of open source projects through pro bono pharma contributions in the future – something I’ve mentioned before. In fact a referee of the OSM paper asked about this, and the wealth of information that arose from one of Derek Lowe’s posts on this subject was then incorporated into the paper – an interesting example of the new synergy between research papers and community discussions.
Pharma’s involvement is a crucial pillar of open source. I have no desire to obliterate pharma, and I’m not sure any other participant in OSM does – there is extraordinary scientific expertise there that can accomplish astonishing things. The experimental inputs to OSM Series 1 from pharma, such as the Rate of Killing Assay, to pick just one example, have been essential for the project. Frederik Deroose from the company Asclepia designed and costed syntheses and agreed to the sharing of these publicly as part of brainstorming processes. The pharma model has weaknesses, meaning they cannot officially pursue certain projects. I think that open source can provide a powerful competing model. But I hope nobody is surprised to see pharma, or indeed the private sector more generally, as co-authors on the paper.
MMV
The Medicines for Malaria Venture (MMV) has tirelessly supported the project. Back in 2010 I gave a talk at the University of Cape Town hosted by Kelly Chibale. Tim Wells happened to be there, and we got to talking about whether the open source idea I’d previously applied to organic synthesis could work in drug discovery. What was interesting about that conversation was how much Tim had thought about this already. We batted around the arguments for and against open source, and soon reached that point familiar to all scientists where you have to acknowledge that discussion can only get you so far – at some point you have to go into the lab and actually do an experiment. We wanted to establish whether open source drug discovery would “work”, i.e. whether a distributed, self-assembled team could work effectively and leverage inputs from the community. Rather than argue the point any more it was going to be simplest just to try it.

I presented the idea to Jeremy Burrows in Geneva, and MMV became more involved. They provided seed funding which got Paul Ylioja to Australia and into the lab to start the very first reaction (10% yield for a Paal-Knorr, mate?). Paul Willis became the day to day champion at MMV. His pharma experience was clear from the outset – a combination of scientific expertise but also a vigorous pursuit of project milestones that was a striking counterbalance to the rather more relaxed set of objectives we find in many academic research projects. MMV have been pioneers in open data for malaria – the best and most recent example being the Malaria Box initiative driven by Paul himself, and the current work around the Pathogen Box.
Early Days in Sydney
MMV’s backing of the project led to us securing a grant from the Australian Research Council, setting up the core of the project in my lab at The University of Sydney. Murray Robertson came on board, and, later on, Alice Williamson. I could not have asked for a more effective and committed set of postdocs to drive the project core, combining synthesis with a huge amount of technical work in setting up how to run an open source project.

Paul Ylioja, Murray Robertson and Alice Williamson
We benefited greatly from the use of the Labtrove Electronic Laboratory Notebook created by Jeremy Frey at The University of Southampton. This allowed the other contributors at Sydney Uni (students Matin Dean, Matt Tarnowski, Zoe Hungerford and Laura White and crystallographer Peter Turner) to share their raw data with the world, and Sanjay Batra and his team at the CDRI in Lucknow (Harikrishna Batchu, Soumya Bhattacharyya) to add their own syntheses.
We really scrambled for solutions in the early days. How best to solicit inputs? How best to encourage inputs from strangers? How best to disseminate results, on what was becoming an aging platform (The Synaptic Leap, created by Ginger Taylor)? How to ensure people did not use email, but instead communicated publicly? How to guarantee permanence of online data and effective linking between posts? (We were ably shepherded through a ridiculous amount of yak shaving by the talented Mike Robins, a neuroscience major at USyd). How to reassure people that raw data in the ELN could indeed be “wrong” but that science can correct itself iteratively and without shame? I remember an event early on that made me think this could all really work: Paul Ylioja received public advice on a troublesome synthesis from someone who clearly had experience of this reaction far beyond the norm – the idea to “age” a reaction overnight after quenching. It worked a treat. We would not have thought of this, and the anecdotal advice came from someone with high levels of technical expertise reading what we were doing as we were failing.
The Community Builds Itself
It’s exactly this feature of openness (experts coming *to* the project spontaneously) that is so powerful.
Everyone has to be on board with the idea of immediate release of all data – one of the contributors swiftest to understand that mission was Corey Nislow, who together with his team (Guri Giaever, Marinella Gebbia, Anna Lee) contributed a great deal of experimental and computational data arising from a yeast-based mechanism of action assay.
Guiding this community and platform development was a growing set of online contributors specialising in informatics. ChEMBL (John Overington, Iain Wallace, George Papadatos) advised the project on how to manage the growing volume of linked data, published the first interim dataset and carried out numerous analyses such as mechanism of action predictions and isosteric replacement strategies described in the paper. Chris Southan kept a watchful eye on patents, and pushed the importance of the discoverability of the molecules in the lab notebooks and other places (here‘s his list of the molecules from Series 1). An advantage of the openness of the lab notebook is that it can be indexed by search engines. Chris schooled everyone in the ways of the SMILEs, the InChi and the InChiKey, allowing people to find the molecules we were working on (ever tried drawing a molecule in the Google search box? Doesn’t work). Chris Swain provided continual advice on how to create, manage and exploit the dataset of all OSM molecules, and Luc Patiny created a way to visualize the structures and their data – for so long in the early days of OSM our poor capability in compound visualisation was a barrier to community participation.
Favourite Moments
Some events stand out.
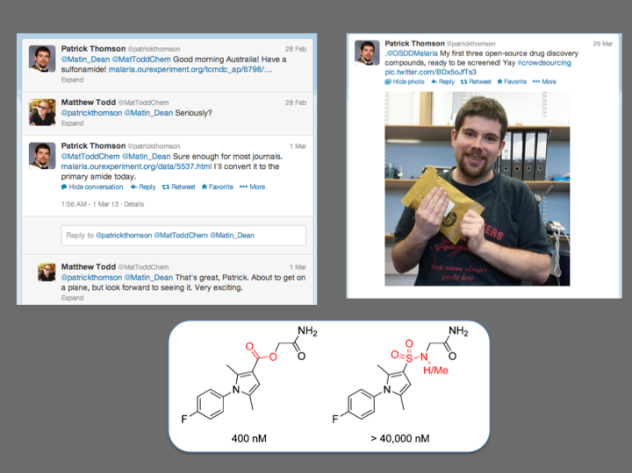
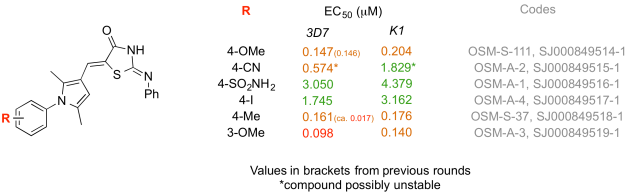
1) Patrick Thomson’s synthesis of the sulfonamide analogue. Patrick, working in Colin Campbell’s lab in Edinburgh, took it upon himself to respond to Alice’s challenge to make the “most wanted” compounds in this series. He communicated with us by Twitter, but all the raw data were on the ELN. He made the molecule faster than we did in Sydney and I won’t forget the day when I woke to find a picture of him on Twitter holding the sample, ready to go. He had it evaluated locally at Dundee (thanks to Irene Hallyburton at the DDU). It was inactive, but an important data point needed for the paper. Patrick continued to make other molecules and contribute to other series.
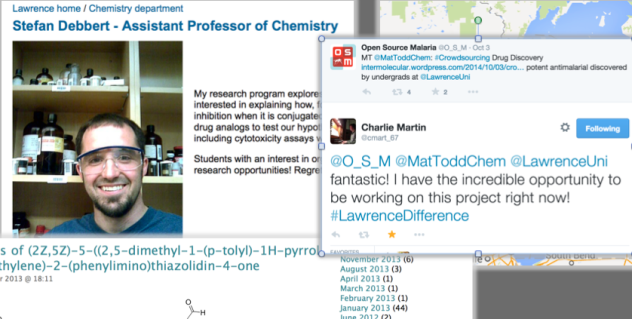
2) Stefan Debbert’s undergrad lab class. Stefan demonstrated the power of crowdsourcing. Open projects work well as a source of undergrad science – all the students can talk about what they are doing and work together on real problems. Stefan managed this with his class, and showed how students can make meaningful contributions to “live” projects. Alice Williamson is now really driving this. That federated lab projects can be run globally, where each lab PI is in total control of the activity but shares data to a core, is a potent idea.
3) Receiving biological data from collaborators and immediately posting the data for community consideration never got old, and remains one of the most exciting aspects of open source. Several leading parasitology labs around the world contributed potency evaluations (e.g. Vicky Avery at Eskitis and her team (Sandra Duffy, Sabine Fletcher), Stuart Ralph at Melbourne Uni and his student James Pham, Kumkum Srivastava in Lucknow, Kip Guy and Julie Clark at St Judes) and evaluations against other parasite stages (Elizabeth Winzeler at UCSD and her team (Stephan Meister, Yevgeniya Antonova-Koch), Michael Delves and Andrea Ruecker at Imperial College). In vivo work, and substantial follow-up analysis was carried out by Sergio Wittlin at Swiss TPHI. In Australia the project benefitted from data and expert guidance from Sue Charman at Monash on solubility and metabolic stability (and her team of Karen White and Eileen Ryan) and Kiaran Kirk’s lab at the ANU (working with his extreme frisbee student Adelaide Dennis) – Kiaran is strongly involved in OSM’s current Series 4, since his favorite antimalarial target, PfATP4, is the target of those compounds. Maybe.
4) The witnessing of productive conversations between professors (Jonathan Baell, who argued strongly for the avoidance of “PAINS”) and undergrads (Zoe Hungerford) to resolve issues quickly within blog posts.
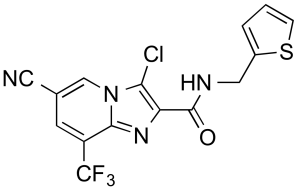
5) The disbelief of witnessing activity cliffs arising from small changes in structure (see the paper itself, Figure 7) – this is something medicinal chemists are all familiar with, but it’s still a shocker when it actually happens to you.
Decision to Park
We communally decided to park the series. It was a tough call – the molecules have striking promise in various ways. If the arylpyrroles had been the last series on the earth, we’d have continued, but there were other series to explore, such as the current in vivo active compounds in Series 4.
But anyone can continue Series 1. There are specific molecules that need to be made, described in the paper, and clear ways forward towards understanding the mechanism of action.
Want In?
The 6th Law of the consortium is about project ownership (see the paper, Figure 1), or rather the lack of it. I started this project with MMV – got it funded, set it up, suggested how it might work best. A core team drove it forwards when it was quiet – open source projects always require leadership. But at heart the OSM consortium is not about ownership, but rather about a way of working. If you cleave to the idea that open data is a good idea, and if you like the idea that when people look over your shoulder they might suggest an improvement to what you’re doing (and that you rise, prospectively, to that challenge) then you’re in. There are no badges, no member dues (there are some T-shirts, actually). Just do an experiment, record the data and expose yourself up to the potential brutality of public improvement.
Other people know this too. The Structural Genomics Consortium is pioneering open data as a stimulus to drug discovery on a large scale. CO-ADD is screening community-contributed compounds for infectious diseases, with a default to public domain data. Sage Bionetworks have pioneered genomics competitions in biomedical research. You can consume your computer’s spare CPU cycles on searching for hits vs. Zika targets using the IBM Grid, or use public data generated by the NIH. I see Open Source Malaria as the necessary extension of open data into the larger, messier place where we all have to roll up our sleeves and make and evaluate real molecules in labs over an extended period.
If you like the idea of Open Source Malaria, then the current series needs work: compound synthesis, biological evaluation and there’s a competition to determine the mechanism of action. There’s also interest in creating an open educational course in medicinal chemistry. And Alice Williamson is keen to work with undergrad lab classes in new analogue synthesis by cloning the course she’s successfully run at The University of Sydney. The current to-do list, a portion of which is shown below, is here. If you have your own series of compounds that you want the community to work on, and you’re happy to share all your data, the platform is for you.
There are varied strands of OSM currently needing inputs
Some Predictions
In poker one can sit around for hours winning or losing little pots of money, but the adrenaline flows in cases where you make significant calls, where you place a lot in the middle on a gut reading of the terrain and then watch it play out. I find long bets interesting, when we’re scoping out where we’re all headed long-term, without enough data. I’m a little unpopular with some for saying that the open access debate is over – open access is in my view the future norm, and there’s nothing anyone can do about it (this is a great thing, incidentally). There is now forming a very significant consensus around open data that will impact us all in the next 2 years (Nature’s new policy, for example). I predict open source to be the norm in scientific research in certain areas in 10 years, and if anyone is working secretively in, for example, neglected disease drug discovery by then they will seem perverse. By the time my kids are adults the idea of secrets in scientific research will probably seem perverse – why hold your team back with secrecy when there are so many advantages in collaborating with the world? I say this because the nature of collaboration is only going one way: to be more seamless, to be faster, to be more machine-enabled. To accommodate this change the legal and economic structures that surround research and research funding are going to need to change, beyond all recognition. The impact of that on the development of new medicines is what I find so exciting. I don’t see a solution that doesn’t have holes, but I’d place a bet that there’s a solution set that will in retrospect seem obvious. And I predict that’ll arise from a proper competition between traditional methods of drug discovery and those that are completely transparent. So many good things in this world come from properly formulated competitions.
FAQs
1. What do you mean by “Open Source”?
All data and ideas freely shared. Anyone can take part. No patents. See the paper, Figure 1.
2. Where is Open Source Malaria?
Landing page (constructed by the talented, responsive people at CloudCity Development), Lab Notebooks, Wiki, Github To Do List, Twitter. Physically located in many places, and contributors come and go.
3. Isn’t this the same thing as Open Source Drug Discovery, in India?
No. OSDD unfortunately shared only a small amount of its activity publicly, so cannot be considered an open source project. OSDD was described more as a crowdsourcing approach to bioinformatics among groups located in India. A malaria project was mooted but did not ultimately receive funding.
4. Is open source the same as “open access”?
No. “Open access” means you can read the results after publication. In open source you can take part in the research before publication.
5. Is open source the same as “open innovation”?
No. “Open innovation” is about securing solutions in new ways, but there is no requirement to collaborate, or to share what you’re doing and there are controls on the release of data.
6. Aren’t you just getting other people to do a bunch of work for free?
No. People contribute voluntarily for any number of reasons. There is no requirement placed on anyone. The consortium is supported by a mix of core funding (government/NGO), funding to contributing labs (e.g. big pharma inputs, or block-funded academic labs doing other things as well), educational projects (student crowdsourcing as part of official courses) and genuine volunteers (who may or may not be working on employer time).
7. You abandoned the series – I guess open source doesn’t work?
Attrition in drug discovery is high. Open source drug discovery is no different on that score. Given all the data are available it’s clear what to do (and what not to do) next for these molecules. In open source there is no unnecessary duplication and no hiding of bad data, meaning if a series fails, it only needs to fail once.
8. Who’s going to pay for the clinical trials for a patentless drug?
There are known ways of doing this, and many more that are more speculative. As yet, there’s no precedent of a open source drug going all the way through to market.
9. Are you all Communists?
No.
























Reply