Open Source Malaria has completed an experiment in crowdsourcing for open drug discovery.
Identifying and developing medicines is a labour-intensive process, particularly in the discovery and optimization phases, and most particularly in the physical preparation of samples of new molecules for testing: a phase that consumes large amounts of time and money and is often a roadblock. One of the obvious things to do is to crowdsource the synthetic chemistry using students.
In open source projects there is the tantalizing additional possibility that student teams can assemble to work on those problems because what’s needed can be fully described in the open. The openness means that the teams could learn from each other, share data and receive peer review and mentorship from interested experts based elsewhere, such as in the pharmaceutical industry.
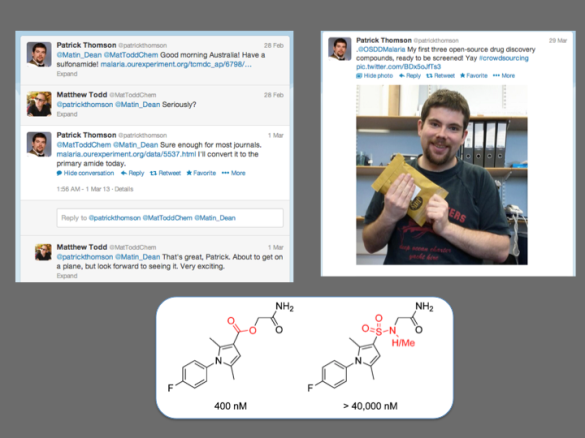
There have obviously been examples of crowdsourcing in various scientific arenas such as genomics (e.g., 1, 2) and there have been closed groups of students operating in the area of drug synthesis. There are examples of crowdsourcing initiatives in attempts to identify biologically active natural products (1, 2), Joerg Bentzien ran a Kaggle competition in in silico small molecule modeling, Urmi Bajpai is working with students in her lab on some biochemical projects (see also this earlier story) and there are other preparative activities of many kinds dating back to things as diverse as the AIDS quilts. I wasn’t aware of any students participating in crowdsourced synthetic chemistry as part of a project that was open source, though Patrick Thomson provided a spectacular example of how openness can lead to high quality scientific contributions from individuals. Mass recruitment of synthetic expertise is going to be one cornerstone of any scaled-up vision for Open Source Pharma, and I was very keen to see if we could complete this exercise in OSM as a precedent, and maybe learn how to make sure it works most effectively.
We just completed this precedent. Around 50 students from Lawrence University in the US midwest worked on the synthesis of six new analogs in the “Near Neighbour” (NN) branch of Series 1 of OSM. The compounds were mailed to a different lab in the US for biological evaluation. Active compounds were discovered – one of them new to the project and quite potent. I find the closure of this loop tremendously exciting.
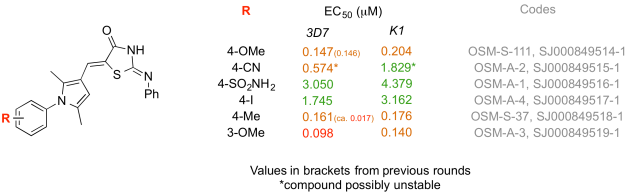
Data for the Lawrence University Compounds – check out the red value for OSM-A-3!
Here’s how it worked. I was contacted a few days before the end of 2012 by two US-based academics asking whether their undergraduate cohorts could contribute to OSM. This had been an aim of the project way back in the design phase and not something we’d gotten round to. For one of the academics, Stefan Debbert, Murray Robertson and I wrote up a description of a synthetic route we’d been using for the NN compounds. The chemistry is not trivial, but possesses the advantage that the compounds are typically solids that can be easily purified and are stable under ambient conditions. Stefan looked into this and came up with some analog structures he felt his students would be able to access, and without any further input from us he got this working in his lab class in early 2013. Around June/July he wrote to say his students had finished the syntheses, and in November he uploaded all the data to the online lab book. After some quality control checking of the data by others, the compounds were shipped to Kip Guy’s lab at St Jude’s where Julie Clark performed the assessment of potency and the data were put up online last month, thereby closing the loop on this part of the project. This whole process took longer than it should have since we were all feeling our way a little on this one.
The data have come in just in time for the writing of the first OSM paper, which is at an advanced stage. We are now faced with the interesting challenge of how to credit this student cohort with authorship. What a great problem to have.

A Student Hard at Work in the Debbert Undergrad Lab
The take-home: a cohort of undergraduates successfully made new molecules that are potent in killing the malaria parasite. The data are valuable as part of a larger project of current research that will be published in the peer-reviewed literature.
The students were able to make an impact because:
1) A current/future research need was openly described
2) The rules behind the project clearly stated that anyone could participate
3) The student team was locally and carefully mentored by a dedicated individual (Stefan) willing to engage in an unusual activity
4) The details of what was needed were described in full, i.e. previous, related research that made it clear the activity would not be open-ended
5) The outputs were actively used, i.e. data deposited in the relevant lab notebook for the benefit of the wider project and molecules tested by a laboratory willing to do so.

This is scaleable. Any team of undergraduates can engage in real research in this way, and perform scientific activities that are valuable to a “live” project and which might result in the undergraduates being able to publish their work. The work could be out-of-hours or, more excitingly (and as in Stefan’s case) part of a formal lab class for which the students receive credit – more on this below. I don’t know about you, but if I’d had the chance to make real molecules for a real research project as an undergrad, I would have found that quite motivating.
One of the neatest aspects about doing this openly is the quality control. A cynical onlooker might say “Well, these are just a bunch of undergrads – how can we trust the work?” You don’t have to. In an open source project, you have access to the raw data, so you can check the quality for yourself. Many people don’t trust data in synthetic chemistry journals in any case (trust is a problem across the discipline) but raw data in open projects solves this problem in as much as it can be solved.
So what would I change next time?
1) Fully Independent Contribution. The collaboration was set up because Stefan contacted me to ask if he could help out. That’s fine, but in an ideal world it would be clear to a lab director what was required without asking – that’s the aim of the Github To Do List that has since been introduced to OSM. But even there it’s sometimes not obvious what’s required and I think we can use Github more effectively. As much as possible in an open project one needs to promote independent contributions and an interlinked mentorship structure (the “Linus doesn’t scale” problem). Having said that, after the initial communications, Stefan ran the whole class independently, which is a testament to his achievement.

The OSM Consortium has a public to do list
2) Direct Data Deposition. The students did not deposit data into the lab notebook directly – Stefan himself aggregated the work and deposited the data for each compound himself. I think this arose because using the lab notebook presented a barrier to participation that was just slightly too large, and there was a concern that students might copy each other. I want to make sure the lab notebook is so easy to use that next time the students are happy to put the data up themselves as they are working – which is the standard practice in OSM. This means making sure that everyone understands that the data they are reading is like any data in any lab book and just because it’s on the internet doesn’t mean it’s correct or final.
One of Stefan’s ELN Entries
3) Global Lab Buddies. Ideally we would have two teams in different places communicating with each other on a synthetic route and not via the central project hub. I have in mind something that ought to be of interest to funding agencies like Fogarty, USAID or Wellcome – a lab class in a developed country pursues a synthetic route and a lab class in a developing nation pursues the same or related experiments, with direct collaboration between students and with mentorship in both places. This has the potential to be an inexpensive way of creating a “lab buddy” scheme where students can share what they learn with each other directly (with only light guidance from a PI, so that the collaboration scales well) and there is no need to spend money moving people around or organizing bench fees; effective lab-based crowdsourcing is significant in terms of how we think about organizing “networks” between universities (e.g. the APRU or WUN) – you can do a lot more for a lot less money if you don’t have to buy airplane tickets.
The experience makes me wonder about best practice:
1) Mentorship. How many mentors are needed for a class? There needs to be a committed local champion like Stefan, and the class size can be left up to the lab director, who also manages local health/safety requirements. There needs to be a central mentor (me, in this case) to help with any snafus. I think there is real potential here for additional mentorship from pharma professionals who could ensure that the molecules being made are reasonable for a drug discovery/development project. Such mentorship could be provided directly pro bono, or organized through a PDP (e.g. MMV, DNDi) or learned organization (e.g. the RSC). Pharma experts in a given location could contribute to their community in this way – e.g. Merck could mentor students working in schools in New Jersey.
2) Replication. It makes sense to me for two students to run each experiment, as a form of replication check. This is real research, so there should be controls, particularly if the students are not very experienced. It’s also probably a good idea for the students to make known compounds as part of the synthetic cluster. The need for positive controls of this kind is particularly keen in open projects since biological evaluation may well be performed in different labs where protocols will inevitably differ. A modest degree of replication makes everyone happier with the numbers ultimately obtained.
3) End-to-End. The work the students do needs to be incorporated into the larger project (at least on a wiki or in a paper) which may require that the molecules are evaluated in some way. If this is needed, then it’s crucial that it happens, so that student effort is not wasted or merely archived. Never ask a crowd to do anything that is not then used. This needs to be factored in to the planning of project resources and means that before the synthesis starts, there needs to be a commitment from somewhere that the molecules will be taken on (in this case, to kill a nasty).
4) How to Assess – or The Brown Oil Problem. I think the greatest power of crowdsourcing lab work lies in incorporating this into the undergraduate curriculum. Imagine that you’re planning a lab class, and you check online to find 38 different current project needs in open malaria, TB and Ebola projects. Let’s say the Open Source Pharma vision is made real and there is a repository of such projects that are known to be active that very day. This is not very far-fetched. You would, I think, be very keen to use one of these real research needs in your class, to fire up your students about how cool research is. If the syntheses were well-designed you could get started as soon as the starting materials were in. But the complication is: How to Assess the Project? What happens if a student fails to prepare a molecule that ought, in theory, to be preparable? How do we assess a project outcome if the outcome is novel, as part of a research project? I have no good answer to this, but feel that there are steps that can be taken:
i) Clarity of Design. The designer of the synthetic route to be undertaken must provide as much detail as possible about the ease of the synthesis. This honesty is simple in open projects – in the NN synthesis undertaken by Stefan it was possible for him to see all of the mis-steps and failures that the OSM team had wrestled with previously, and it was therefore possible for us to provide a lot of advice about key steps. Students themselves can even cite previous attempts as mitigating circumstances (“Though the molecule could not be prepared it was noted that Mat Todd similarly failed to generate this key intermediate in experiment 34-6 (13th Oct 2012)…” etc)
ii) Emphasis on Approach. The marking scheme used by the lab director needs to focus more on the approach used by the student and the quality of the scientific record produced than on the final outcome. This is in any case good academic practice. There remains an issue of plagiarism if students are posting items into the public domain, but one has to wonder about the value of having the students write reports that are vulnerable to this in the modern age.
iii) Time-resistance. The marking scheme needs to be immune to later discoveries. If it is ultimately found that a particular compound is unstable, a student who has earlier made this compound and found the same phenomenon cannot have their efforts re-marked, just because this makes the assessment too conditional and complex.

OSM is supported financially and scientifically by the Medicines for Malaria Venture and the Australian Government
If people want to get involved in this kind of activity, and it’s not clear from the OSM To Do List or the wiki what is needed, then don’t hesitate to get in touch directly. It’d be great to generate some more synthetic schemes suitable for students. If you are planning a research project yourselves, and are writing a proposal, consider including some element of crowdsourcing+open source so we can have lots more students contributing to real research in the public domain.










Reply